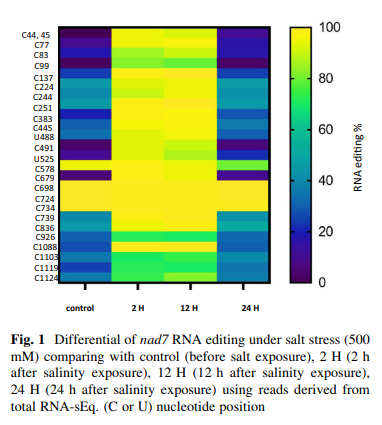
Salinity stress reveals three types of RNA editing sites in mitochondrial Nad7 gene of wild barley both in silico and in qRT-PCR experiments
Cellular respiration is an important process performed by mitochondria. Nad complex is the major complex involved in this process and one of the main subunits in this complex is the nad7 (nad dehydrogenase subunit 7). In Hordeum vulgare subsp. spontaneum, four nad7 cDNAs are described at 500 mM salinity, 0 h, or control (GenBank accession no. MW433884), after 2 h (GenBank accession no. MW433885), after 12 h (GenBank accession no. MW433886) and after 24 h (GenBank accession no. MW433887). Twenty six RNA editing sites were revealed in positions: C44, C45, C77, C83, C99, C137, C224, C244, C251, C383, C445, U488, C491, U525, C578, C679, C698, C724, C734, C739, C836, C926, C1088, C1103, C1119, and C1124. These alterations represent three types of editing: C to U, U to C and the very rare type U to G with a differential rate depending on exposure times. Two and twelve hours are shown as the maximum editing rate. However, RNA editing disturbance over 24 h was observed, possibly due to the activation of the machinery for programmed cell death. In addition, RNA editing has been found to optimize codon bias by altering the nucleotides without modifying the amino acids. Small kernel 1 pentatricopeptide repeat protein (Smk1) characterization in wild barley has assisted to explaining the behavior of C836 editing under salt stress. The behavior of nad7 editing under stress may lead to a discrepancy between the editing of RNA and the control of cellular respiration. © 2021, The Author(s), under exclusive licence to Brazilian Society of Plant Physiology.