Benchmarking of Antimicrobial Resistance Gene Detection Tools in Assembled Bacterial Whole Genomes
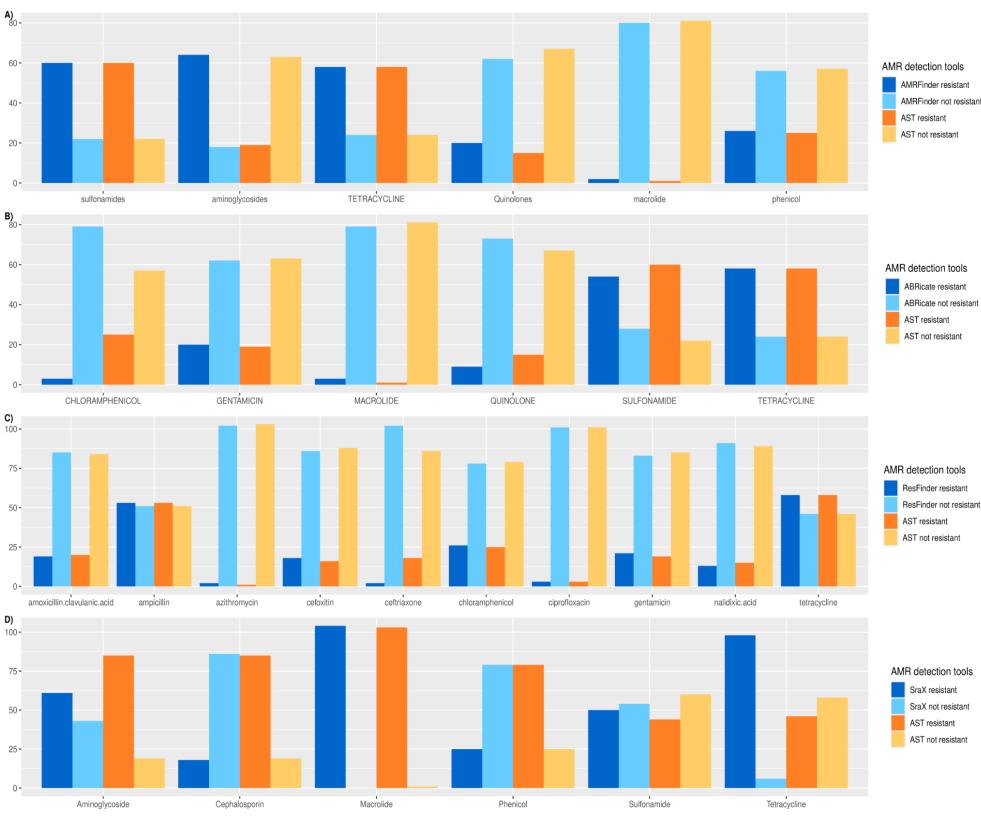
Antimicrobial resistance (AMR) is one of the ten dangers threatening our world, according to the world health organization (WHO). Nowadays, there are plenty of electronic microbial genomics and metagenomics data records that represent host-associated microbiomes. These data introduce new insights and a comprehensive understanding of the current antibiotic resistance threats and the upcoming resistance outbreak. Many bioinformatics tools have been developed to detect the AMR genes based on different annotated databases of bacterial whole genome sequences (WGS). The number and structure of databases used may affect prediction quality. Herein, we aim to check the performance of four AMR gene detection tools and characterize the detection quality by comparing predicted results to reference antibiotic susceptibility test (AST) data. This may enhance the precise in-silico prediction of resistance phenotype and reduce false-positive predictions, which lead to more biologically relevant results. Four AMR gene detection tools; AMRFinder, ABRicate, ResFinder, and SraX are used for the benchmarking using Salmonella enterica isolates (n=104) retrieved from National Center for Biotechnology Information (NCBI) Assembly Database in July 2021. Performance is checked in terms of accuracy, precision, and specificity for each tool. Pearson χ2 test is used to compare predicted results with antibiotic susceptibility testing (true results). All performance measures are assessed via scikit-learn package 0.21.3 and R software V 4.0.3. The highest accuracy was achieved by AMRFinder (0.89), while ResFinder had the highest precision score (0.93) and ABRicate has the lowest time and memory consumption. On the other hand, ResFinder's results confirmed the null hypothesis of the Pearson χ2 test. We conclude that ResFinder is the best tool where its results have the tiniest difference compared to the phenotypic antibiotic susceptibility (true results). © 2021 IEEE.