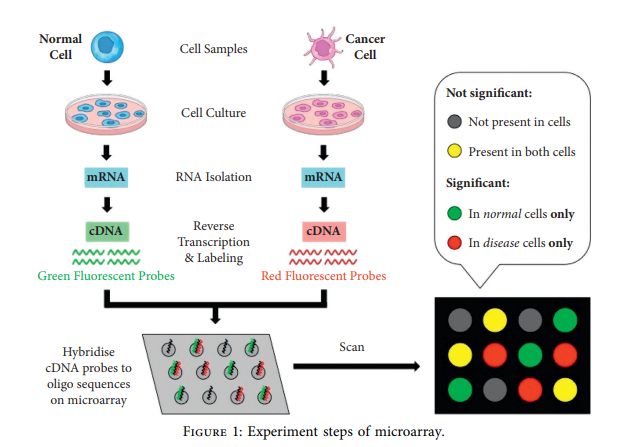
An Efficient Cancer Classification Model Using Microarray and High-Dimensional Data
Cancer can be considered as one of the leading causes of death widely. One of the most effective tools to be able to handle cancer diagnosis, prognosis, and treatment is by using expression profiling technique which is based on microarray gene. For each data point (sample), gene data expression usually receives tens of thousands of genes. As a result, this data is large-scale, high-dimensional, and highly redundant. The classification of gene expression profiles is considered to be a (NP)-Hard problem. Feature (gene) selection is one of the most effective methods to handle this problem. A hybrid cancer classification approach is presented in this paper, and several machine learning techniques were used in the hybrid model: Pearson's correlation coefficient as a correlation-based feature selector and reducer, a Decision Tree classifier that is easy to interpret and does not require a parameter, and Grid Search CV (cross-validation) to optimize the maximum depth hyperparameter. Seven standard microarray cancer datasets are used to evaluate our model. To identify which features are the most informative and relative using the proposed model, various performance measurements are employed, including classification accuracy, specificity, sensitivity, F1-score, and AUC. The suggested strategy greatly decreases the number of genes required for classification, selects the most informative features, and increases classification accuracy, according to the results. © 2021 Hanaa Fathi et al.